Areas of Study
Epigenetics can be broadly defined as the study of heritable changes in organisms caused by modification of their gene expression rather than alteration of the genetic code itself. This definition is still tenuous as "heritable" can mean mitotic or meiotic depending on the parties discussing the matter. For my work we will assume the more loose mitotic definition, although our work on DNA methylation has the possibility of meiotic transmission.
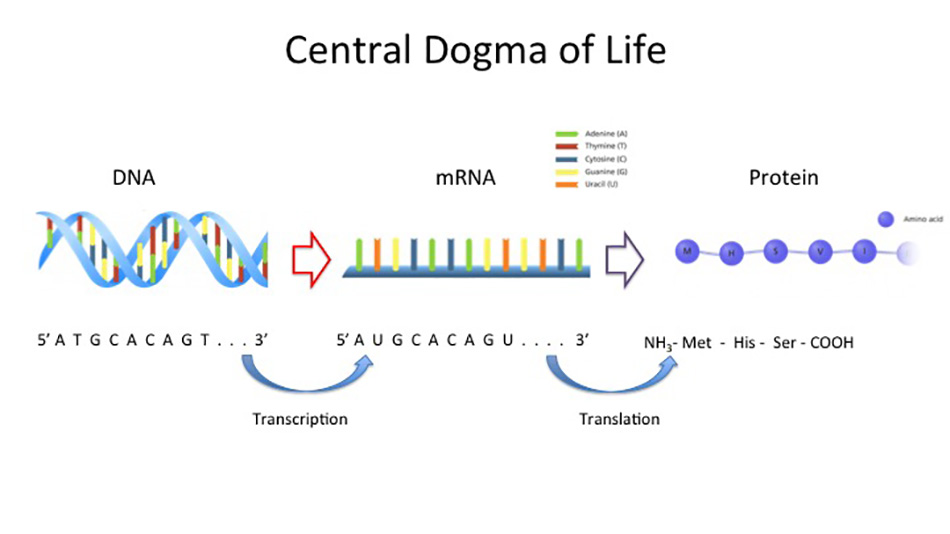
Throughout evolution life has developed a diverse repertoire of mechanisms to control the expression of genes, which are the functional units of DNA. All forms of life utilize an information flow pipeline where genes are transcribed to mRNA which are translated into protein. Although the flow of information from blueprint to message to machine is fascinating at all steps, I focus on the question of how molecular life achieves precise control of the message making process. To describe gene regulation in a musical analogy: if genes are notes in a song, then the regulation controlling when and how long those musical notes are played is what allows a song to be more than a non-sensical mess. Correct regulation of gene expression is what allows the symphony that is cellular life to faithfully carry out developmental programs in the face of many environmental obstacles. It is this regulation that scientists speak of when they discuss epigenetics.
Ignoring epigenetic controls, transcription of the DNA blueprint into messenger RNA (mRNA) requires a large number of nuclear proteins including RNA Polymermase II, transcription initiation factors, elongation factors, mediators, splicing proteins and generalized transcription factors that respond to environmental and developmental cues. Going a level deeper we find that the entire transcriptional process previously described can be either permitted or blocked via multiple passive and active mechanisms. Epigenetic mechanisms that control chromatin status include: recruitment/blocking of transcription factors during binding gene promoters, control of local chromatin compaction, control of high order chromatin compaction (interaction among genome elements that are spatially close in the nucleus).
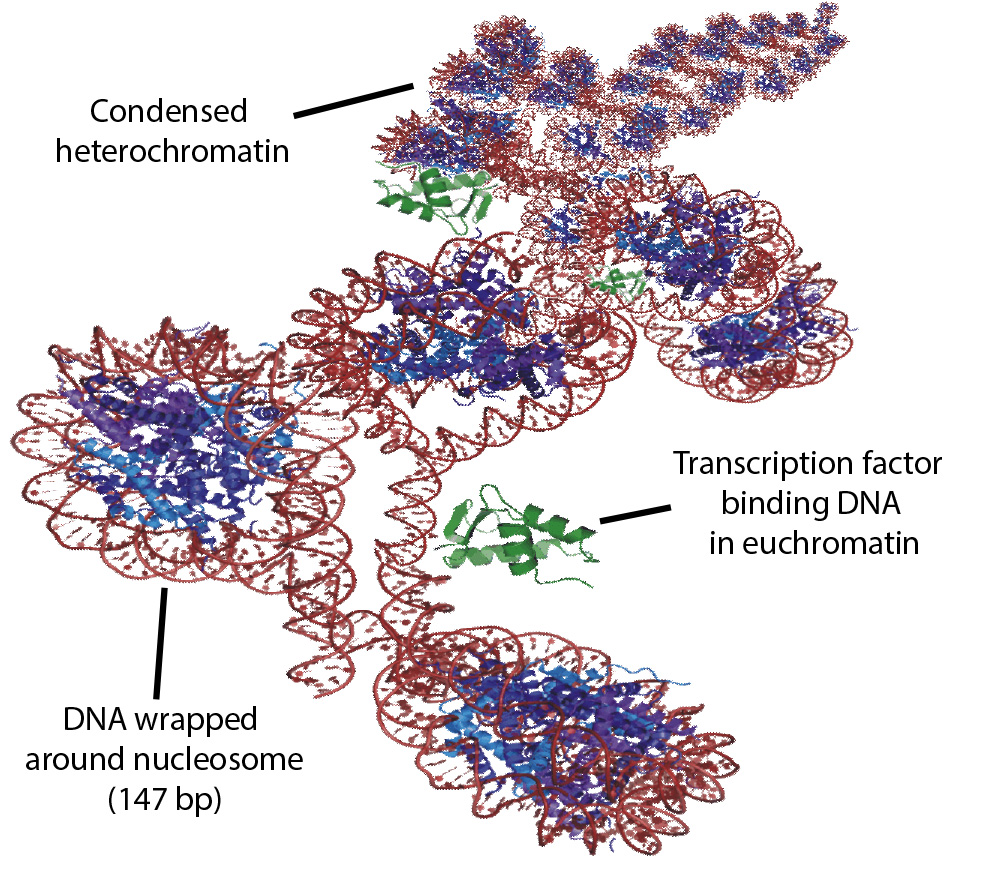
My research is concerned with charecterizing how chromatin state (euchromatin (permissive) or heterochromatin (intolerant) of transcription) affects gene expression related to stress response in plants. Because of their sessile nature, plants must adapt to abiotic stressors quickly to avoid death. Indeed experiments have shown that plants that are given low dose abiotic stresses survive better when they are later given a stronger stress treatment. In the cases where a plant survives a stress episode it may be possible to pass on the memory of an abiotic stress episode via epigenetic mechanisms to further generations. However, transgenerational inheritance of stress tolerance (where epigenetic mechanisms would be required), is still heavily debated in all manner of eukaryotic organisms.
My current project focuses on DNA methylation. We've found that specific DNA methylation mutants in plants have differential survival during heat stress. Because these mutations lead to gains and loses of DNA methylation at specific areas of the genome we came to the conclusion that DNA methylation is likely important to heat stress tolerance.
This observation lead us to ask multiple questions which we are still testing experimentally:
1. Where is the DNA methylation changing in these mutants and how does that affect local gene expression?
2. Does DNA methylation status alter local chromatin compaction?
3. How does DNA methylation interact with histone modifications during heat stress?
4. Does DNA methylation change lead to changes in intra- and inter-generational stress memory?
To test questions 1-3 we are utilizing multiple high throughput genomic techniques (RNA, Bisulphite, ATAC and ChIP-sequencing) and Tandem Mass Spectrometry (for Histone Modifications) on stressed and non-stressed plants. By combining temporal data on DNA methylation, gene expression, histone modification, and chromatin accessibility collected from leaf tissue we hope to determine how DNA methylation alterations lead to phenotypic consequences in abiotic stress. We'll then utilize genetics to confirm the DNA methylation regulated genes that are essential to stress tolerance. After determining which genes are influenced by DNA methylation we will perform inter-generational stress experiments and utilize our previous data to determine if epignenetic mechanisms can lead to transgenerational tolerance.
